Accelerating Python Code with Numba to C++-like Speeds
1. Introduction
Numba is a Just-in-Time (JIT) compiler for Python. This means that when you call a Python function, all or part of your code is converted “on-the-fly” into machine code, which then runs at the speed of your native machine! It is sponsored by Anaconda, Inc. and supported by many other organizations.
With Numba’s help, you can accelerate all computationally intensive Python functions (e.g., loops). It also supports the NumPy library! So, you can use NumPy in your computations and speed up overall calculations, as loops in Python are notoriously slow. You can also use many functions from Python’s standard math library, such as sqrt, etc.
2. Why Choose Numba?

So, why choose Numba when there are many other compilers like Cython and PyPy? The reason is simple: you don’t have to leave the comfort zone of writing Python code. That’s right, you don’t need to change your code at all to get a speedup comparable to what you’d get from similar type-defined Cython code. Isn’t that great?
You just need to add a familiar Python feature: a wrapper (a decorator) to your function. Class decorators are also under development.
So, you just need to add a decorator, for example:
from numba import jit
This still looks like native Python code, doesn’t it?
3. How to Use Numba?

“question mark neon signage” by Emily Morter on Unsplash
Numba uses the LLVM compiler infrastructure to convert native Python code into optimized machine code. Code run with Numba can achieve speeds comparable to similar code in C/C++ or Fortran.
Here’s how the code is compiled:
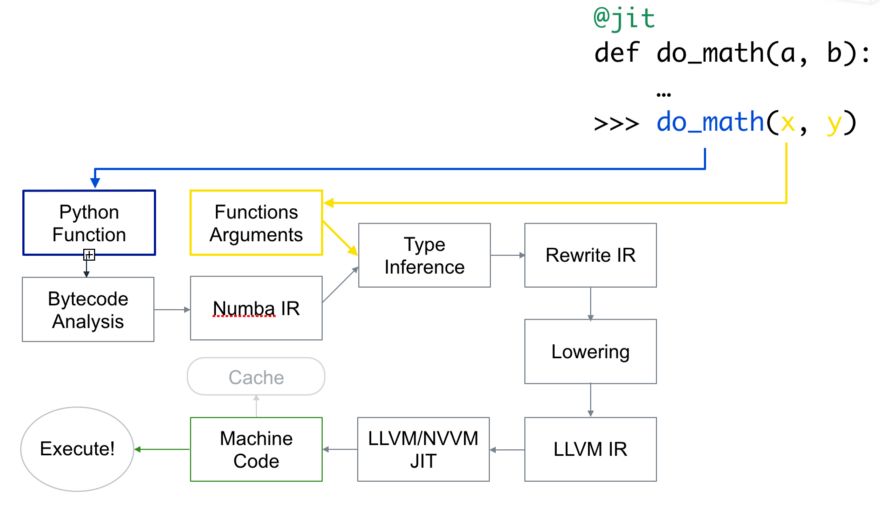
First, the Python function is passed in, optimized, and converted into Numba’s intermediate representation. Then, after type inference (similar to NumPy’s type inference, where a Python float becomes a float64), it’s converted into LLVM-interpretable code. This code is then fed to LLVM’s JIT compiler to generate machine code.
You can generate machine code at runtime or at import time as needed. Compilation at import time can occur on the CPU (default) or GPU.
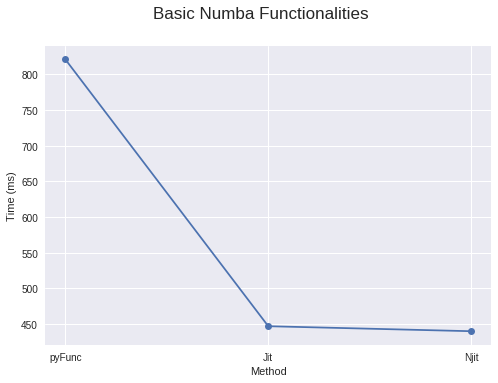
4. Basic Numba Usage (Just Add @jit!)

Photo by Charles Etoroma on Unsplash
Piece of cake!
For optimal performance, Numba actually recommends adding the nopython=True parameter to your jit decorator. This prevents the use of the Python interpreter. Alternatively, you can use @njit. If your decorator with nopython=True fails with an error, you can use the simpler @jit decorator to compile parts of your code. For code it can compile, it will convert them into functions and compile them into machine code. The remaining parts of the code will then be handed over to the Python interpreter.
So, you just need to do this:
from numba import njit, jit
When using @jit, ensure your code contains something Numba can compile, such as computationally intensive loops involving libraries (like NumPy) and functions it supports. Otherwise, it won’t compile anything, and your code will run slower than without Numba due to the extra overhead of Numba’s internal code checks.
Even better, Numba caches functions after their first use as machine code. So, it will be faster after the first use because it doesn’t need to recompile the code if you’re using the same parameter types as before.
If your code is parallelizable, you can also pass parallel=True as an argument, but it must be used with nopython=True. Currently, this only works for CPU.
You can also specify the function signature you want your function to have, but then it will not compile for any other parameter types you provide. For example:
from numba import jit, int32
Now your function can only receive two int32 type arguments and return an int32 type value. This way, you have better control over your function. You can even pass multiple function signatures if needed.
You can also use other decorators provided by Numba:
@vectorize: Allows scalar arguments to be used as NumPy ufuncs.@guvectorize: Generates NumPy generalized ufuncs.@stencil: Defines a function to be a kernel function for stencil-type operations.@jitclass: Used for JIT compilation of classes.@cfunc: Declares a function for native callbacks (to be called by C/C++ etc.).@overload: Registers your own function implementation for use in nopython mode, e.g.:@overload(scipy.special.j0).
Numba also features Ahead-of-Time (AOT) compilation, which generates compiled extension modules that do not depend on Numba. However:
- It only allows regular functions (ufuncs are not supported).
- You must specify the function signature. And you can only specify one signature; if multiple signatures are needed, you must use different names.
It also generates generic code based on your CPU architecture family.
5. @vectorize Decorator

“gray solar panel lot” by American Public Power Association on Unsplash
By using the @vectorize decorator, you can transform functions that operate only on scalars (e.g., if you’re using Python’s math library which only works for scalars) so that they can be applied to arrays. This provides speed similar to NumPy array operations (ufuncs). For example:
@vectorize
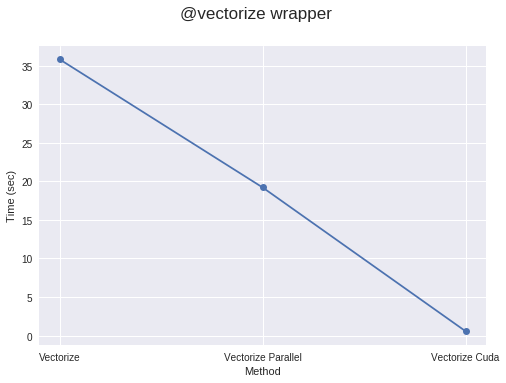
You can also pass a target parameter to this decorator. Setting target='parallel' enables parallelization for the code, while target='cuda' makes it run on CUDA/GPU.
Using @vectorize(target='parallel') or 'cuda' for vectorization often runs faster than NumPy-implemented code, provided your code is computationally dense enough or the arrays are sufficiently large. If not, it might take longer due to the overhead of creating threads and distributing elements across different threads. Therefore, the computational workload should be large enough to achieve a noticeable speedup.
This video demonstrates an example of using Numba to accelerate the Navier-Stokes equations for computational fluid dynamics:
6. Running Functions on GPU

“time-lapsed of street lights” by Marc Sendra martorell on Unsplash
You can also pass @jit as a decorator to run functions on CUDA/GPU. For this, you must import cuda from the numba library. However, running code on the GPU is not as straightforward as before. To run functions on hundreds or even thousands of threads on the GPU, some initial calculations are required. In fact, you must declare and manage the hierarchy of grids, blocks, and threads. This isn’t that difficult.
To execute functions on the GPU, you must define a function called a kernel function or a device function. Let’s first look at kernel functions.
Some key points to remember about kernel functions:
a) A kernel function must explicitly declare its thread hierarchy when called, i.e., the number of blocks and the number of threads per block. You can compile a kernel function once and then call it multiple times with different block and grid sizes.
b) Kernel functions have no return value. Therefore, either changes must be made to the original array, or another array must be passed to store the results. To compute scalars, you must pass single-element arrays.
So, to launch a kernel function, you must pass two arguments:
- Number of threads per block.
- Number of blocks.
For example:
threadsperblock = 32
The kernel function in each thread must know which thread it is in to understand which elements of the array it is responsible for. Numba makes it easy to get the positions of these elements with a single call.
@cuda.jit
To save time wasted copying NumPy arrays to the specified device and then storing the results back into NumPy arrays, Numba provides several functions to declare and send arrays to the specified device, such as numba.cuda.device_array, numba.cuda.device_array_like, numba.cuda.to_device, etc., to avoid unnecessary copying back to the CPU (unless necessary).
On the other hand, device functions can only be called from within the device (by kernel functions or other device functions). A good point is that you can return values from device functions.
from numba import cuda
You should also check out the features supported by Numba’s cuda library here.
Numba also includes its own atomic operations, random number generators, shared memory implementations (to speed up data access), and other features in its cuda library.
ctypes/cffi/cython interoperability:
cffi: Supports callingCFFIfunctions in nopython mode.ctypes: Supports callingctypeswrapped functions in nopython mode.- Cython exported functions are callable.
Conclusion
This article primarily introduced how to use Numba to accelerate Python code execution speed. It can be helpful when performance is a requirement.
- 原文作者:春江暮客
- 原文链接:https://www.bobobk.com/en/313.html
- 版权声明:本作品采用知识共享署名-非商业性使用-禁止演绎 4.0 国际许可协议进行许可,非商业转载请注明出处(作者,原文链接),商业转载请联系作者获得授权。